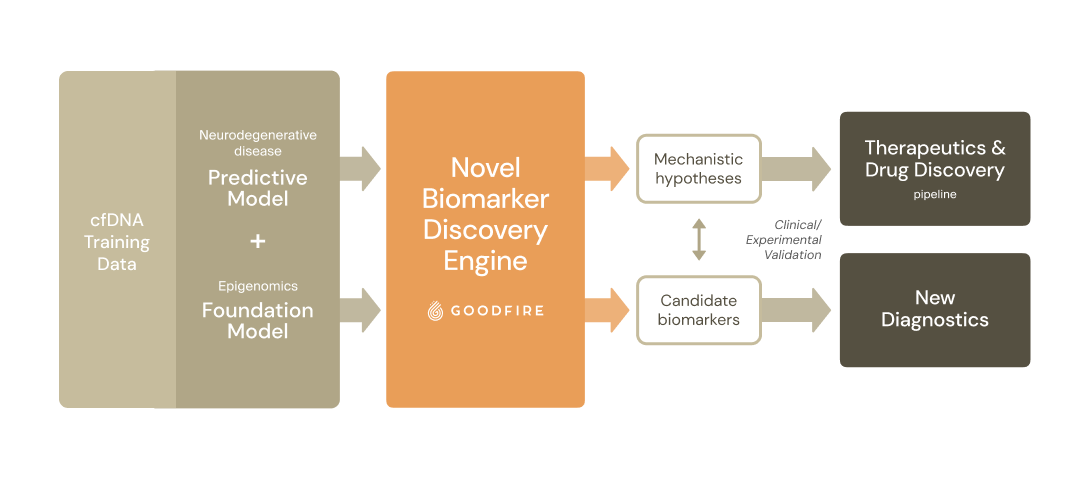
Prima Mente’s epigenomics model had state-of-the-art performance for early detection of Alzheimer’s Disease - but in order to experimentally validate specific biomarkers and move towards FDA approval, they needed to narrow down a huge number of possible signals their model might have been using. Goodfire’s platform for in silico science decoded their model, identifying a novel class of biomarkers for Alzheimer’s detection while providing insights for improving the model’s design.
Context
Prima Mente is an AI neuroscience company working across the entire AI-driven discovery cycle.
They’ve trained a series of foundation models on the human epigenome, operate a wet lab to validate hypotheses generated by their model, and run clinical trials for new therapies. Their Pleiades model series was trained on 1.9 trillion tokens of raw human epigenomic data in order to understand neurodegenerative disease, pushing state-of-the-art performance in detecting Alzheimer’s disease from a single blood sample.
Outcomes
Prima Mente, together with Goodfire, identified a novel class of blood-borne biomarkers for Alzheimer’s detection.
If validated, these results pave the way for clinical applications in minimally invasive diagnosis of neurodegenerative disease and help identify promising targets for the development of therapeutics. These biomarkers are currently undergoing experimental validation and will be detailed in a forthcoming publication.
The Challenge
Prima Mente’s model had high accuracy for detecting neurodegenerative disease, but they couldn’t identify what signals their black-box model was actually using. They needed to:
- Decide which hypotheses to spend valuable wet lab hours on and deploy to the clinic for diagnostics, choosing from thousands of possible biomarkers
- Extract the model’s understanding of disease mechanisms to develop targets for therapeutics
- Jumpstart improvement and iteration on their model design by getting signal on how their model works
The model presented several challenges to interpretability:
- limited data size, confounders, and uncertainty in which signals would generalize to new patients
- a black-box model with no natural language interface or interpretable features
- heterogeneous, overlapping inputs differing in cell type, region, sampling method, and even the per-patient epigenome itself
- a complex hierarchical model architecture which aggregates many sources of information, making it hard to trace signals to individual inputs
Our Approach
Prima Mente partnered with Goodfire to understand its epigenomics model. Goodfire’s interpretability platform, tool suite, and infrastructure - coupled with its expertise in both interpretability and AI for scientific discovery - turned their foundation model into an engine for biomarker discovery.
Goodfire’s research scientists embedded in Prima Mente’s team as they had finished training their model, and built out a biomarker discovery pipeline:
- trained sparse autoencoders (SAEs) on Prima Mente’s model to extract meaningful intermediate features
- traced predictions back through the model to specific, interpretable signals in the data
- tested the identified signals’ robustness and generalization to new patients
- ablated the primary signals to understand subtler contributions masked by dominant features
For more details on the approach and results, see our research post.